Search
About Anopheles merus
Range
Anopheles merus belongs to the Anopheles gambiae species complex, which consists of at least seven species, and it is a locally important vector in eastern and southern Africa where it is mainly found along the coast.
Habitats
An. merus is found in high numbers in shallow brackish pools and marsh or swamp areas along the coast. As a consequence, this species does not exhibit density changes in response to tidal fluctuations nor does it appear to tolerate very high levels of salinity. It is rarely found in the mangrove forests on the east coast of Africa, however this may be due to the composition of the trees and soil type under of the stands of mangrove in this zone rather than inherent behavioural patterns. An. merus is known to occur inland, using salt pans and saline pools as larval habitats.
Resting and feeding preferences
An. merus is generally opportunistic in host selection, depending on host availability and females have a tendency to bite and rest outdoors. The biting times of An. merus on the Kenyan coast have been reported as gradually rising from early evening (18:00) peaking between midnight and 01:00 and then declining until 06:00, which reflects the accepted biting pattern for this species across its range.
Vectorial capacity
An. merus has previously been considered as only a minor or unimportant vector, however, it has been identified as playing an unexpectedly important role along the Tanzanian coast and more recently in Mozambique.
This text was modified from Sinka ME et al. (2010) The dominant Anopheles vectors of human malaria in Africa, Europe and the Middle East: occurrence data, distribution maps and bionomic précis Parasites & Vectors 3:117.
MAF strain
Originally isolated from South Africa (Mafayeni, Kruger National Park), isofemale selection was performed prior to genome sequencing. This strain is available from BEI resources.
Source: VectorBase
Picture credit: James Gathany, CDC Public domain via Wikimedia Commons (Image source)
Taxonomy ID 30066
Data source VectorBase
Variation
What can I find? Short sequence variants.
 More about variation in Ensembl Metazoa
More about variation in Ensembl Metazoa




 Download DNA sequence
Download DNA sequence Convert your data to AmerM2 coordinates
Convert your data to AmerM2 coordinates Display your data in Ensembl Metazoa
Display your data in Ensembl Metazoa
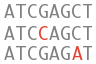

![Follow us on Twitter! [twitter logo]](/i/twitter.png)
