Search
NOTE: The genome sequence and annotation for T. muris are updated in Ensembl Metazoa infrequently. The data here corresponds with WormBase WS279. For the latest genome and annotation, please visit WormBase ParaSite (for an Ensembl-oriented view), or WormBase.
About Trichuris muris
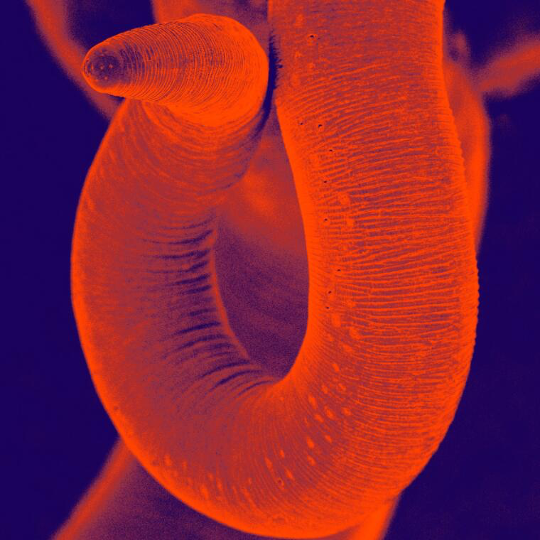
The nematode Trichuris muris, or murine whipworm, is a gastrointestinal parasite of mice which resides in the caecum and colon and has a direct oral faecal life cycle. It is a remarkably tractable model system for dissecting immune responses and host parasite relationships and is actively being investigated in a number of laboratories worldwide.
Photo credit: David Goulding, Wellcome Trust Sanger Institute Attribution-NonCommercial 4.0
Taxonomy ID 70415
Data source WormBase
Comparative genomics
What can I find? Homologues, gene trees, and whole genome alignments across multiple species.
 More about comparative analyses
More about comparative analyses
 Phylogenetic overview of gene families
Phylogenetic overview of gene families
 Download alignments (EMF)
Download alignments (EMF)
Variation
This species currently has no variation database. However you can process your own variants using the Variant Effect Predictor:



 Display your data in Ensembl Metazoa
Display your data in Ensembl Metazoa
 Update your old Ensembl IDs
Update your old Ensembl IDs

![Follow us on Twitter! [twitter logo]](/i/twitter.png)
