Trichuris muris (Nematode) Assembly and Gene Annotation
NOTE: The genome sequence and annotation for T. muris are updated in Ensembl Metazoa infrequently. The data here corresponds with WormBase WS279. For the latest genome and annotation, please visit WormBase ParaSite (for an Ensembl-oriented view), or WormBase.
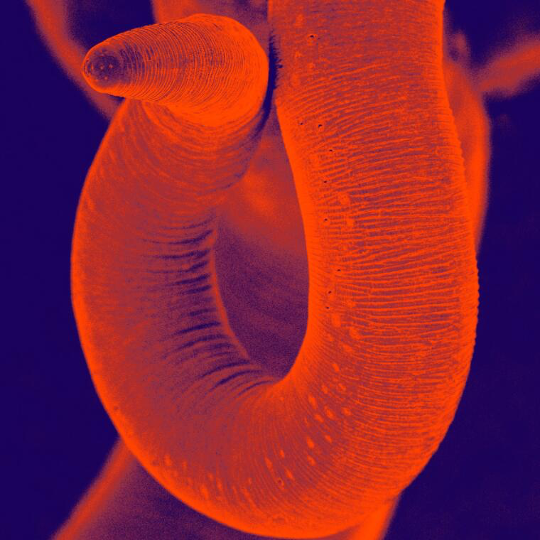
About Trichuris muris
The nematode Trichuris muris, or murine whipworm, is a gastrointestinal parasite of mice which resides in the caecum and colon and has a direct oral faecal life cycle. It is a remarkably tractable model system for dissecting immune responses and host parasite relationships and is actively being investigated in a number of laboratories worldwide.
Photo credit: David Goulding, Wellcome Trust Sanger Institute Attribution-NonCommercial 4.0
Assembly
The T.muris 2.x reference genome has been sequenced by the Berriman Lab in collaboration with Richard Grencis, University of Manchester, as described by Foth et al (2014). The 3.x reference assembly is an improvement adding more sequencing data by the Berriman Lab.
Annotation
The original gene predictions were made by the Parasite Genomics group at the Wellcome Trust Sanger Institute, as described by Foth et al (2014). A new geneset has been created for the 3.x assembly by WormBase.
References
- Whipworm genome and dual-species transcriptome analyses provide molecular insights into an intimate host-parasite interaction.
Foth BJ, Tsai IJ, Reid AJ, Bancroft AJ, Nichol S, Tracey A, Holroyd N, Cotton JA, Stanley EJ, Zarowiecki M, Liu JZ, Huckvale T, Cooper PJ, Grencis RK, Berriman M. 2014. .Nat Genet, 2014;46(7):693-700
Picture credit: File:Trichuris_muris image provided by David Goulding, Wellcome Trust Sanger Institute. This file is licensed under the Attribution-NonCommercial 4.0 International (CC BY-NC 4.0) license and be used for any purpose, as long as it is not primarily intended for or directed to commercial advantage or monetary compensation.
Links
Statistics
Summary
| Assembly | TMUE3.0, INSDC Assembly GCA_000612645.2, |
| Database version | 111.279 |
| Golden Path Length | 111,837,642 |
| Genebuild by | WormBase |
| Genebuild method | Import |
| Data source | WormBase |
Gene counts
| Coding genes | 14,995 |
| Non coding genes | 759 |
| Small non coding genes | 759 |
| Gene transcripts | 18,200 |



![Follow us on Twitter! [twitter logo]](/i/twitter.png)
